Intro to Reproducible Science
Adapted from the Reproducible Science Curriculum
Special Thanks: Francois Michonneau, Hilmar Lapp, Karen Cranston, Jenny Bryan, and everyone else who contributed to these materials.
NEON has adapted them to our week long Data Institute.
Reproducibilty is actually all about being as lazy as possible!
– Hadley Wickham (via Twitter, 2015-05-03)
Why Use Reproducible Methods?
More efficient, less redundant science: others can build upon our work.
Reproducibility & Your research
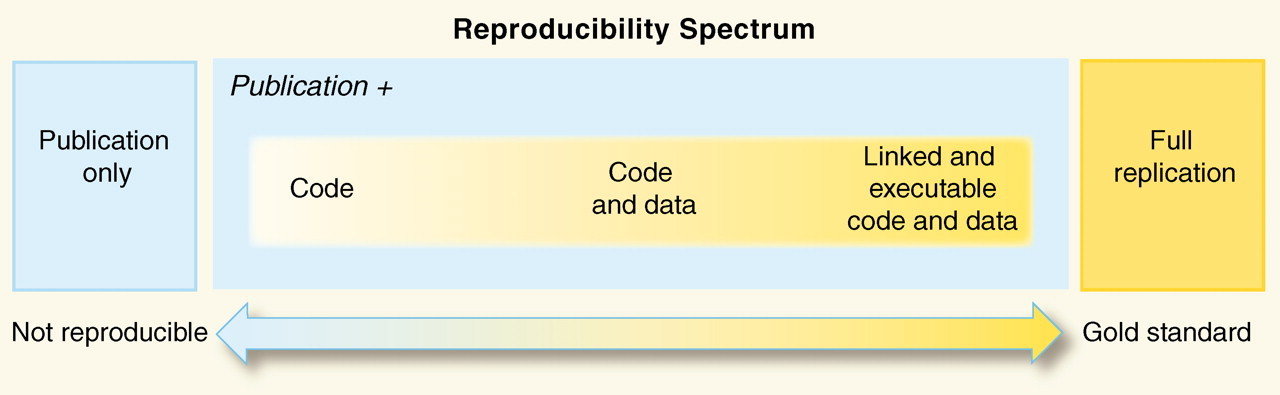
Reproducibility spectrum for published research. Source: Peng, RD Reproducible Research in Computational Science Science (2011): 1226–1227 via Reproducible Science Curriculum
Five selfish reasons to work reproducibly - Florian Markowetz
Reproducibility helps to avoid disaster
Reproducibility makes it easier to write papers
Reproducibility helps reviewers see it your way
Reproducibility enables continuity of your work
Reproducibility helps to build your reputation
How to Make Work Reproducible
For research to be reproducible, the research products (data, code) need to be publicly available in a form that people can find and understand them.
Who do we need to share with?
Collaborators
Peer reviewers & journal editors
Broad scientific community
The public
Better Research
Figure 1. Distribution of reporting errors per paper. Papers from which data were shared has fewer errors.
Click on citation to view paper.
The tools that make this easier
GitHub: Version Control / Collaboration / Dissemination
R Markdown or Jupyter Notebooks: Code Documentation / Dissemination
Four Facets of Reproducibility
Over the next week, we will focus on the tools and skills associated with these facets.
- Organization
- Automation
- Documentation
- Dissemination
1. Organization
The more self explanatory the better:
- Consider overall structure of folders and files
- Use informative file names
1. Organization Pro-Tip
A variable name that describes the object is more useful than a random variable name.

Noble, William Stafford, 2009. A quick guide to organizing computational biology projects.
1. Organization
File Organization should:
- Reflect inputs, outputs and information flow
- Preserve raw data so it’s not modified
- Carefully document & store intermediate & end outputs
- Carefully document & store data processing scripts
Organization
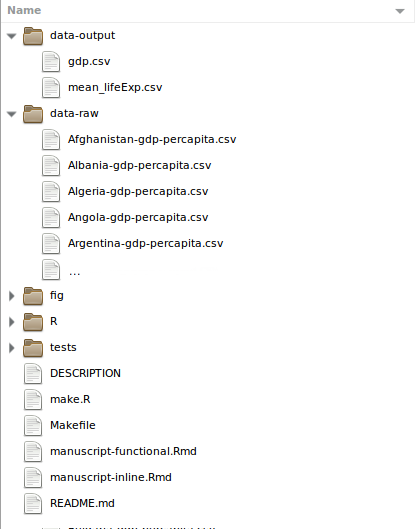
1. Organization – File Names
File / Folder Names should be:
- Machine readable
- Human readable
- Support sorting
Which set of file names are most self-explanatory?
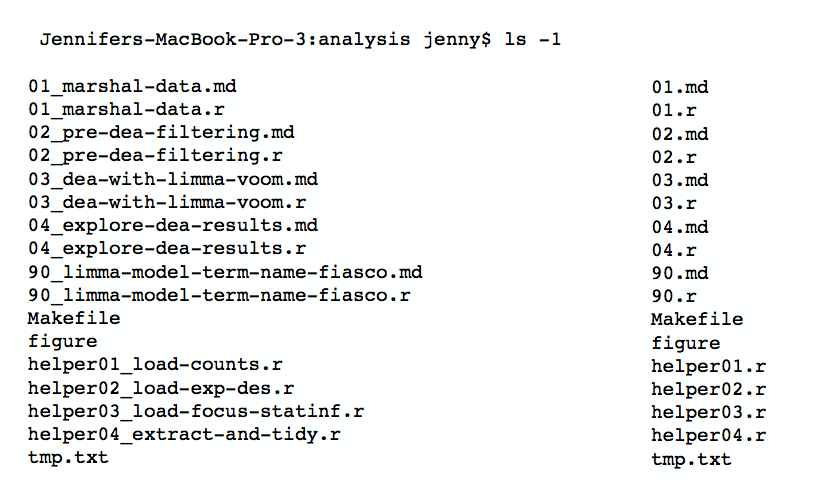
More on file naming & organization
– from the Reproducible Science Curriculum
1. Organization - Benefits
- Your future self will be able to quickly find files
- Colleagues will be able to more quickly understand your workflow
- Machine readable names can be quickly and easily sorted and parsed
2. Automation Pro-Tip
Scripting vs. Point and click
Script = more time spent up front, but will save time in the long run.
2. Automation Pro-Tip
Time Savings:
- More efficient to modify and repeat an analysis down the road
- Easier for reviewers and colleagues to see even aspect of your methods
- Self documenting methods - your future self will forget small steps
2. Automation
DRY – Don’t Repeat Yourself
If your analysis is composed of scripts, with repeated code throughout, it will be more time consuming to maintain and update.
Automation Tips
Modularity – use functions to write code in reusable chunks
- Variables created within a function are temporary
- Code with functions can be easier to read / cleaner
- Allows for better documentation
- Supports testing
- Allows for re-use of code on other data
2. Documentation
Document all workflow steps:
- You can remind your future self of your workflow
- Others can see and understand your work
- Future “re-analysis” of your data is more efficient
Documentation
Code should be easy to understand with clear goals
Document your code even if you think it’s clear and simple. Your collaborators & your future self will inevitably have an easier time working with it down the road.
Documentation Pro-Tip 1
Add comments around functions that describe purpose, inputs and outputs.
Documentation Pro-Tip 2
Avoid proprietary formats: Use text files (.txt, .md) that don’t require special tools to open.
Documentation Pro-Tip 3
Markdown to style documentation = machine readable, small file size, low overhead.
Documentation Pro-Tip 4
Use coding approaches that connect data cleaning, analysis & results
R Markdown and Jupyter notebooks allow you to publish code and results in one (or more) output files.
Dissemination
Publishing is not the end of your analysis, rather it is a way towards your future research and the future research of others.
Dissemination - Why
- Funding agency / journal requirement
- Community expects it
- Increased visibility / citation
- More efficient, less redundant science
Dissemination workflow
Example Workflow / Tools:
Document workflow: R Markdown / Jupyter Notebooks
Collaborate with Colleagues / Version Control : GitHub
Publish Data Snapshot: FigShare, Dryad, Zenodo, etc
Share workflow: Notebook Viewer, Binder
Four Facets of Reproducibility
An overview of some of the topics, tools and skills that we will cover during the Data Institute
Documentation: Jupyter notebooks, GitHub
Organization: File naming / directory structure best practices
Automation: Efficient Coding Practices (in Python)
Dissemination: GitHub
Questions?
Email: neondataskills@BattelleEcology.org